
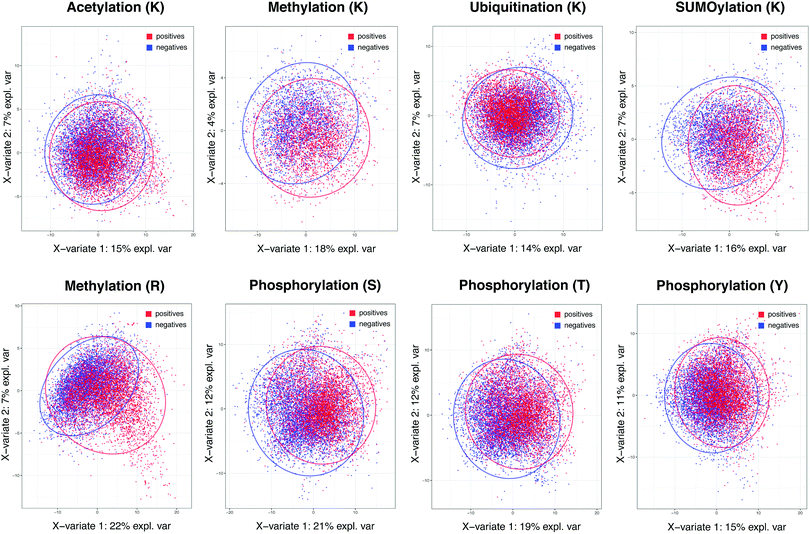
The reader gets the details of both experimental and computational methods and strategies in the identifications and functional annotation of proteins.
This edition includes recent cutting-edge technologies and methods for protein identification and quantification using tandem MS techniques. In addition, specific MS/MS spectrum similarity scoring functions and their application in the field of proteomics, statistical evaluation of labeled comparative proteomics using permutation testing, and methods of phylogenetic analysis using MS data are also described in detail. The most recent proteomic resources widely used in the biomedical scientific community for storage and dissemination of data are discussed. This book covers the most recent proteomics techniques, databases, bioinformatics tools, and computational approaches that are used for the identification and functional annotation of proteins and their structure. Thus, MS-based proteomics continuously improved the way proteins are identified and functionally characterized. Preface Recently, mass spectrometry (MS) instrumentation and computational tools have witnessed significant advancements. Printed on acid-free paper This Humana Press imprint is published by Springer Nature The registered company is Springer Science+Business Media LLC The registered company address is: 233 Spring Street, New York, NY 10013, U.S.A. Neither the publisher nor the authors or the editors give a warranty, express or implied, with respect to the material contained herein or for any errors or omissions that may have been made. The publisher, the authors and the editors are safe to assume that the advice and information in this book are believed to be true and accurate at the date of publication.
Peptideshaker ptm color free#
in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. The use of general descriptive names, registered names, trademarks, service marks, etc.

All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. Suresh Mathivanan Department of Biochemistry and Genetics La Trobe Institute for Molecular Science La Trobe University Melbourne, VIC, Australia Shivakumar Keerthikumar and Suresh Mathivanan Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC, AustraliaĮditors Shivakumar Keerthikumar Department of Biochemistry and Genetics La Trobe Institute for Molecular Science La Trobe University Melbourne, VIC, Australia Walker School of Life and Medical Sciences University of Hertfordshire Hatfield, Hertfordshire, AL10 9AB, UK Shivakumar Keerthikumar Suresh Mathivanan Editors


 0 kommentar(er)
0 kommentar(er)
